COVID-19 All Research
IARPA research efforts assisting the Intelligence Community and United States to combat SAR-CoV-2
BioHEAT
Identifying Critical Viral Targets for Improved Vaccines and Therapeutics Development
BioHEAT is a short duration, limited scope effort to develop new methods for analyzing the stability and relative mutation rate of proteins. The program is creating a visual representation of evolutionarily stable regions and mutation hotspots within multiple SAR-CoV-2 proteins across coronavirus families and strains from the current pandemic, which may be used for better contact tracing.
April 30, 2020: Baylor researchers develop new modeling techniques to understand genetic stability of SARS-CoV-2, enabling faster discovery of therapeutics and vaccines
Baylor College of Medicine, an IARPA performing team, is researching a novel Evolutionary Trace and Evolutionary Action software pipeline to analyze the stability and relative mutation rate of proteins. The team, in consultation with the National Biodefense Analysis and Countermeasures Center, used the pipeline to compare publicly available reference sequences and create a visual representation of evolutionarily stable regions and mutation hotspots within multiple SAR-CoV-2 proteins across coronavirus families and strains from the current pandemic. The analysis also validated previous studies that indicated mutations present appear to be natural in origin. This new computational pipeline enables tracking of mutations in the viral genome across time and space, potentially enabling improved contact tracing. Additionally, identification of stable regions may be used to develop medical countermeasures and high specificity detection platforms.
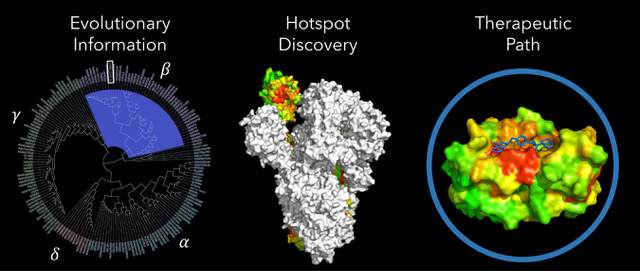
BioHEAT COVID-19 research: This seedling identifies functional hotspots in proteins and whether they arose through natural evolutionary processes or by unnatural engineering. Integrative analytics efficiently exploit evolutionary information, here in the Coronavirus superfamily (left). This produces a structural map of a natural functional hotspot in the Spike proteins (middle). A hotspot can serve as an antigenic site, a lead for peptide mimetic drugs, or, here, a target for docking repurposed drugs or discovery of new drug formulations. A comprehensive map of hotspots in all SARS-CoV-2 proteins is under development. (Figure used with permission)
Crowdsourcing Evidence, Argumentation, Thinking and Evaluation (CREATE)
Crowdsourcing Platform Facilitates Development of Policy Recommendations for COVID-19 Recovery
IARPA’s CREATE program developed tools and methods designed to improve analytic reasoning through the use of crowdsourcing and structured analytic techniques. These resources empower multi-disciplinary collaboration among analysts. A software platform developed under CREATE has been used to enable a geographically dispersed team of researchers to work together to analyze COVID-19 policy options related to social distancing.
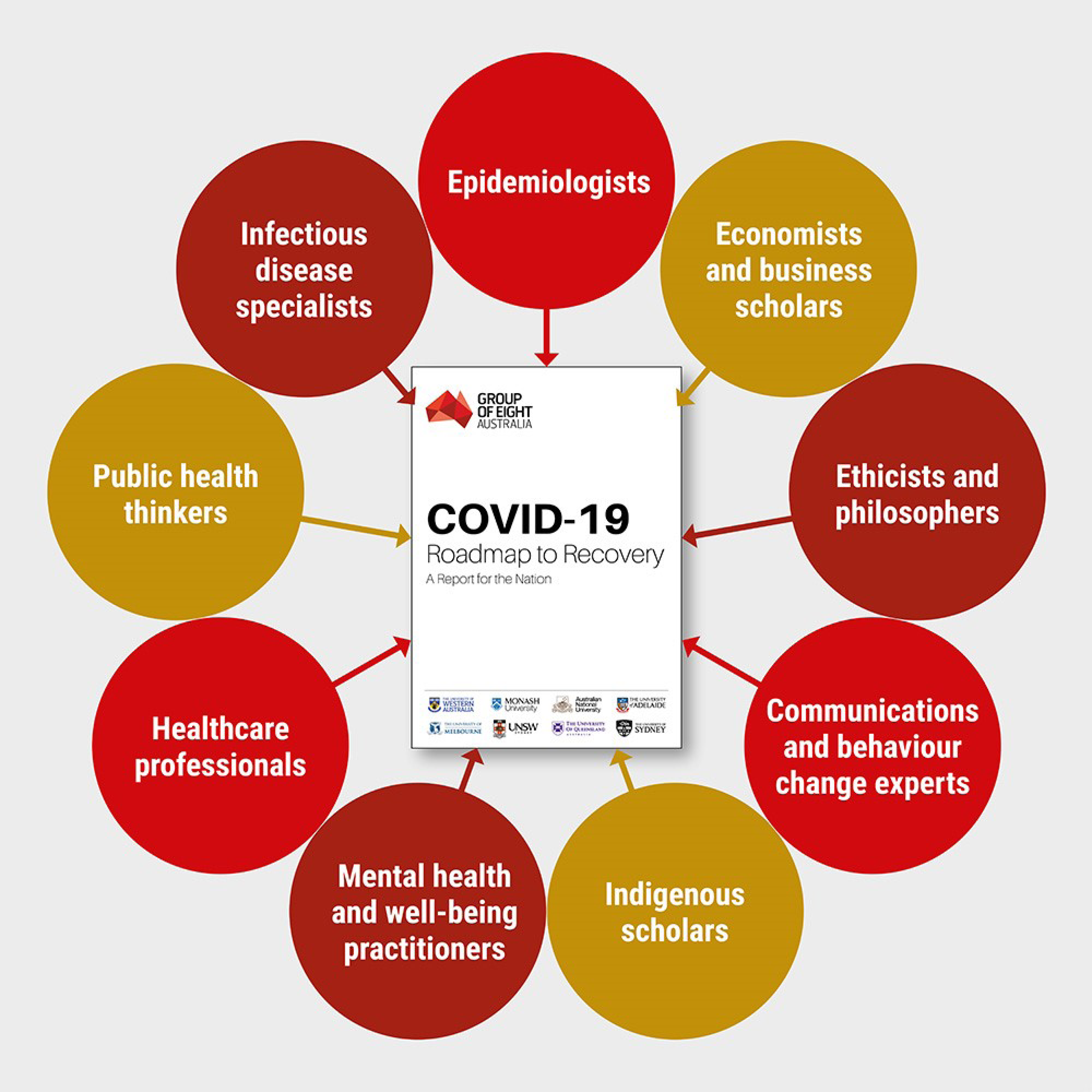
April 18, 2020: IARPA-developed collaboration tool enables Australian researchers to build a Roadmap to Recovery during the COVID-19 outbreak
An article in the April 18, 2020 issue of The Australian described a collaboration among roughly 100 researchers from Australia’s eight leading universities to analyze possible outcomes of policy decisions to relax COVID-19 social distancing restrictions. The university taskforce used a collaborative software platform developed under IARPA’s CREATE program. The software platform, the Smartly-assembled Wiki-style Argument Marshalling project, facilitates the aggregation of high volumes of complex information, analysis of consequences, and packaging of recommendations into a succinct report. For three weeks, the taskforce examined a range of questions ranging from whether there should be uniform approaches across Australia, rules for travel and border protection, preparations for reopening schools and businesses, special consideration for the elderly, rural residents, indigenous people, and citizens needing mental health support, and communications strategies to encourage public support and cooperation. The report, entitled the Roadmap to Recovery, was delivered to the Australian Health Minister and the Chief Medical Officer at the end of the April.
Finding Engineering-Linked Indicators (FELIX)
Rapid Evaluation of SARS-CoV-2 For Signatures of Engineering
Under the scope of the FELIX program, IARPA is developing new computational and experimental tools to rapidly detect indicators of genome engineering in multiple types of biological systems, including viruses, improving and augmenting existing detection capabilities. Designed to work across a range of biological organisms and sample types, such as environmental collections with a mixture of organisms, the FELIX tools and methods can provide early alerts to the presence of engineered organisms and help expedite appropriate responses thereby avoiding adverse consequences.
January 2020: IARPA’s FELIX Program Investigates Rumors that COVID-19 Pandemic is the Result of Genetic Engineering
The MIT-Broad Foundry, a performer team on the FELIX program, analyzed the publicly available SARS-CoV-2 genome using their FELIX bioinformatics pipeline in order to test the veracity of online stories claiming that SARS-CoV-2 was engineered in a laboratory. They compared the SARS-CoV-2 genome against 58 million sequences, including genomes from closely and distantly related viruses. After only 10 minutes of analysis, the FELIX tool determined that all regions of the SARS-CoV-2 genome match naturally-occurring coronaviruses better than they match any other organisms, including any other viruses. This analysis indicates that no sequences from foreign species have been engineered into SARS-CoV-2.
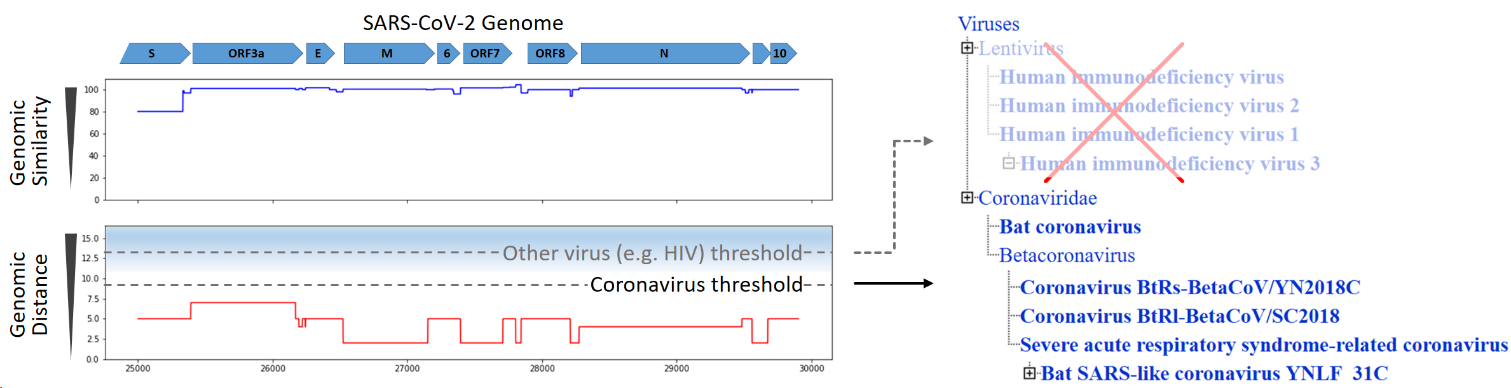
Figure 1: Comparison of the SARS-CoV-2 genome to a comprehensive sequence database shows that the closest genetic matches are to other coronaviruses. Image used with permission.
Functional Genomic and Computational Assessment of Threats (FunGCAT)
Revealing the Mysteries of How SARS-CoV-2 Disrupts Our Immune System
The Fun GCAT program is substituting current simplistic look-up table-based methods to flag dangerous DNA sequences with smart, AI-driven threat screening software that considers DNA function, can process short or highly engineered sequences, and does not require follow-up analysis by an expert. Fun GCAT is also uncovering the cellular and molecular roles of DNA, sequence by sequence, using new experimental pipelines. One pipeline is testing virus genes for the ability to disrupt the immune system hardwired within each human cell.
June 25, 2020: Raytheon BBN Technologies applies new DNA screening software to accelerate the development of better SARS-CoV-2 diagnostics
Raytheon BBN Technologies is applying its FAST-NA software, funded under the IARPA Fun GCAT program, to rapidly screen for DNA sequences unique to the SARS-CoV-2 virus. The goal is a low-cost, point-of-care diagnostic test that will use saliva and a paper strip to deliver test results for SARS-CoV-2 in less than an hour. FAST-NA was originally developed for screening commercial DNA synthesis orders by defining and flagging unique DNA signatures of pathogenicity. The software identifies the specific DNA sequences associated with SARS-CoV-2 for the diagnostic assay to detect the virus, with confidence, in human saliva. The SARS-CoV-2 diagnostic test development is a collaboration with Purdue University, PortaScience Inc., Coretex Design Inc., and LaDica LLC.
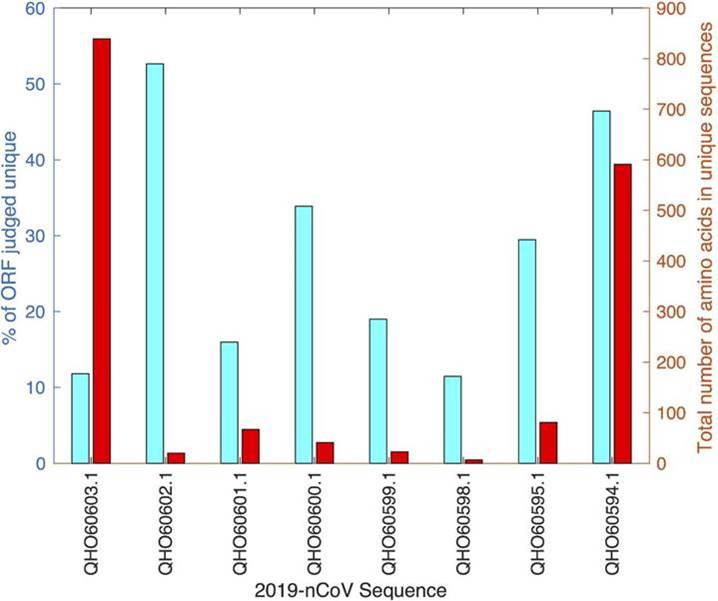
Summary statistics of distinguishing amino acid sequences for SARS-CoV-2 compared to all other Coronaviridae genomes isolated using the FAST-NA software. For each open reading frame (ORF) indicated on the x-axis, the fraction of the ORF judged to be unique and the total number of amino acids in unique sequences is presented. As an example, the large 1ab polyprotein, QHO60603.1, has many unique residues, though the fraction is not large. In contrast, the surface glycoprotein, QHO60594.1, has both a large amount and large fraction of unique material. The unique region of the surface glycoprotein, identified in January, has since been determined to be a critical region of the spike protein tied to the infectiousness of COVID-19. The same technology is now being used to identify stable diagnostic targets. Image Credit: Jacob Beal, Thomas Mitchell, Daniel Wyschogrod, Jeff Manthey, and Adam Clore (doi: https://doi.org/10.1101/2020.01.31.929497)
February 2020: The Fun GCAT Program and Harvard University are exploring how SARS-CoV-2 evades and disrupts the human immune system
Researchers at Harvard University led by Professor Pamela Silver recently developed an experimental pipeline for the evaluation of viral genes that disrupt the human immune response as part of IARPA’s Functional Genomic and Computational Assessment of Threats program. This pipeline is specifically tailored to the rapid analysis of novel viruses during the early stages of an outbreak from engineered cell lines (Fig 1A). With the outbreak of COVID-19, the Harvard team has shifted priorities to analyze and compare a first set of 120 genes from SARS-CoV-2, the virus that causes COVID-19, and six closely related coronaviruses (e.g., SARS, MERS, and more common circulating variants) to reveal how SARS-CoV-2 disrupts our immune system and what genetic features make the virus particularly pathogenic (Fig 1B). An improved understanding of coronavirus immune evasion mechanisms is essential for the development of future vaccines and therapeutics. While these types of investigations can take months of dedicated work, the Fun GCAT team at Harvard has developed rapid analysis approaches that take only days from sequence receipt to answer, significantly accelerating our capabilities.
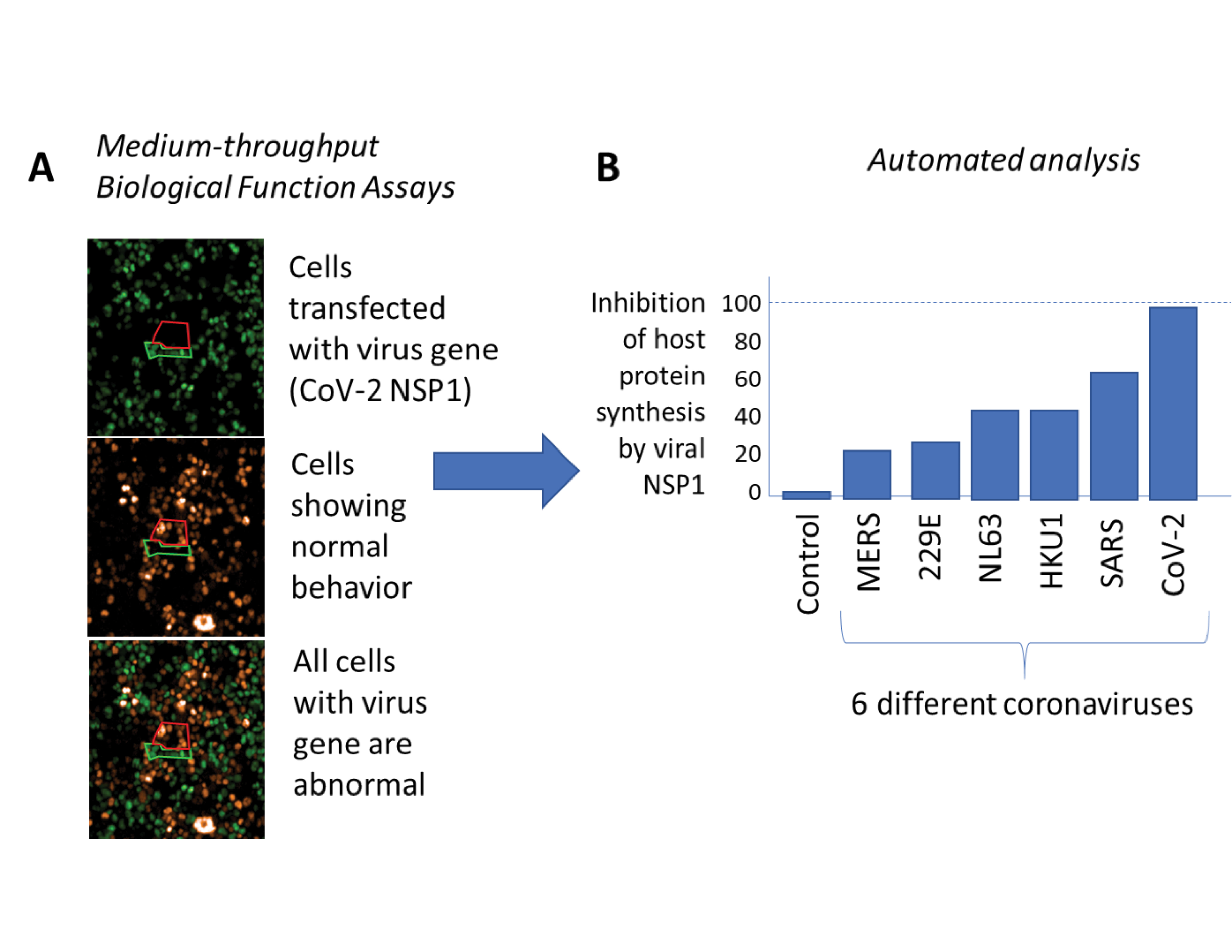
Figure 1: High-throughput testing of many coronavirus genes reveals that the NSP1 gene of the SARS-CoV-2 virus is the most powerful of the genes tested at inhibiting protein synthesis and the antiviral response of the infected cell. Credit: Timothy Chang, Erika Olson, Jeffrey Way, and Pamela Silver. (Figure used with permission)
Molecular Analyzer for Efficient Gas-phase Low-power INterrogation (MAEGLIN)
Early Breath-Based Detection of SARS-CoV-2 Related Respiratory Disease
The MAEGLIN program has been developing sensors to detect harmful gaseous chemicals in air. The goal is detection of weapons of mass destruction (WMD) or chemical indicators of illicit activity (e.g., narcotics production). In a COVID-19 pivot, MAEGLIN is now investigating the program’s new micro-gas chromatograph as a breath sensor to detect signs of Acute Respiratory Distress Syndrome (ARDS), a life-threatening condition associated with COVID-19.
April 28,2020: IARPA MAEGLIN Program Sensor Starts Clinical Trials
Researchers have completed assembly of the first test device (Fig. 1) and received final approvals for the clinical trial protocols. Initial results of the clinical trial are expected be available in June 2020, with more complete results available later in the summer.

Figure 1: The sensor being used in clinical trials is packaged in a shock-resistant and easily decontaminated carrying case. A ventilator or breathing tube can be attached directly to the sensor intake. A laptop connected via micro-USB collects data with the flip of a switch without requiring an expert to operate the device. (Photo used with permission.)
April 3, 2020: The MAEGLIN Program and University of Michigan pivot towards the development of a non-invasive, breath-based sensor for pre-clinical detection of ARDS
As part of IARPA’s Molecular Analyzer for Efficient Gas-phase Low-power INterrogation program, the University of Michigan developed a micro-gas chromatography (µ-GC) system (Fig. 2) to sample and identify hazardous chemicals in the air. In preliminary clinical trials, the sensor proved capable of diagnosing diseases such as ARDS and asthma from exhaled breath (Fig. 3). Preliminary testing prior to the COVID-19 outbreak showed the µ-GC was able to identify patients who would later show ARDS symptoms more than 24 hours earlier than the existing clinical diagnostics. Early identification of patients likely to suffer from ARDS could enable earlier treatment of the large fraction of COVID-19 patients who develop this life-threatening condition associated with the virus. The µ-GC system is undergoing an accelerated clinical trial to assess its effectiveness as a non-invasive diagnostic tool, to help triage patients and better allocate resources, to help monitor COVID-19 patients through the recovery phase, to inform treatment and identify potential relapses. IARPA expects the first devices to be delivered in May 2020 and clinical trials to begin soon thereafter.
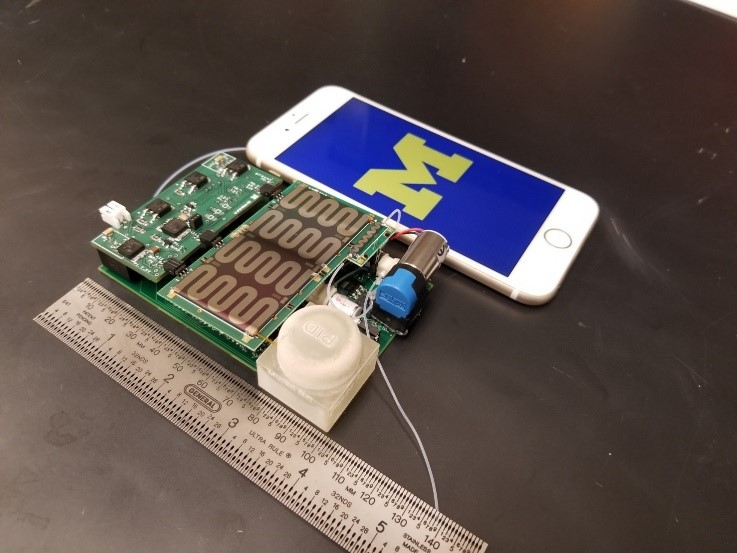
Figure 2: Sensor platform developed by the University of Michigan in Phase 1 of the MAEGLIN program. This is nearly identical to the sensor being tested in the clinical trials. (Photo used with permission.)
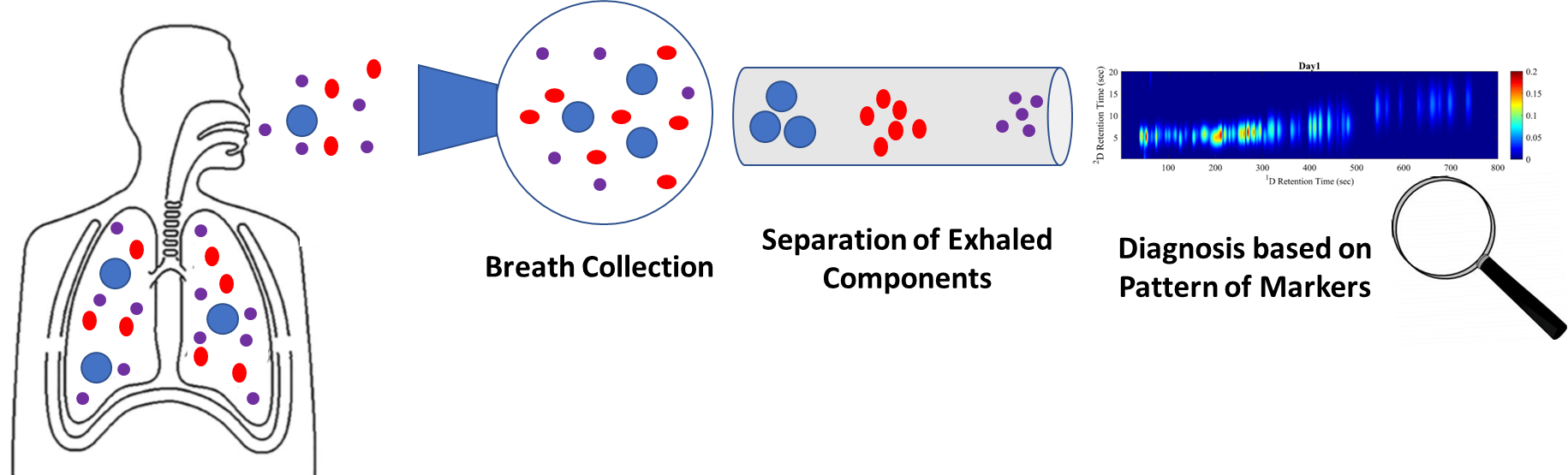
Figure 3: Proposed use of the MAEGLIN University of Michigan µ-GC as a breath sensor for medical conditions.
IC Postdoctoral Research Fellowship Program
The Intelligence Community Postdoctoral Research Fellowship Program was established in 2000 to support unclassified basic research in areas of interest to the IC. Funded primarily by the Office of the Director of National Intelligence, the program annually supports several postdoctoral fellows from U.S. accredited colleges, universities, and U.S. government laboratories across the country.
Intelligence Community Research on Audio Algorithms Pivots to Support Emergency Ventilator Efforts During COVID-19 Response
Dr. Ryan Corey is an IC Postdoctoral Fellow at the University of Illinois at Urbana-Champaign supported under an IARPA sponsored topic on research algorithms for distributed audio networks. Recently, Ryan made a major contribution to the development of a monitoring system for emergency ventilators. Many low-cost ventilators developed to address the COVID-19 pandemic lack the electronic monitoring and alarm features found in commercial ventilators. Dr. Corey and colleagues adapted a signal processing algorithm used to control loudness in hearing aids to efficiently process pressure signals measured in a ventilator to alert clinicians when specific errors occur. The Illinois RapidAlarm team designed the monitoring system to complement the Illinois RapidVent, a low-cost emergency ventilator that has been licensed by several manufacturers worldwide. The designs and source code for the Illinois RapidAlarm system are freely available online. The device can be constructed quickly at low cost using widely available parts, helping address medical supply shortages with safer, more useful emergency ventilators
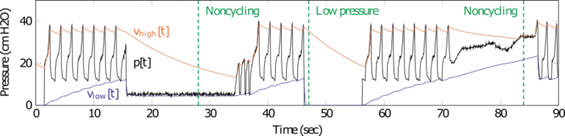
Figure 1: Simulated alarm conditions initiating a response by the algorithm
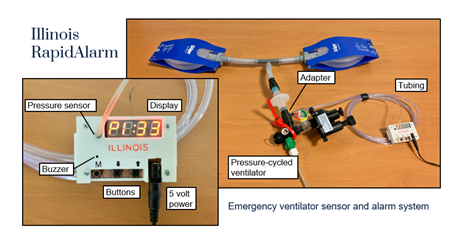
Figure 2: Prototype of Illinois RapidAlarm sensor and alarm system attached
